Cell Reproduction
10.1 – The Process of Meiosis
Learning Objectives
By the end of this section, you will be able to do the following:
- Describe the behavior of chromosomes during meiosis, and the differences between the first and second meiotic divisions
- Describe the cellular events that take place during meiosis
- Explain the differences between meiosis and mitosis
- Explain the mechanisms within the meiotic process that produce genetic variation among the haploid gametes
Sexual reproduction requires the union of two specialized cells, called gametes, each of which contains one set of chromosomes. When gametes unite, they form a zygote, or fertilized egg that contains two sets of chromosomes. (Note: Cells that contain one set of chromosomes are called haploid; cells containing two sets of chromosomes are called diploid.) If the reproductive cycle is to continue for any sexually reproducing species, then the diploid cell must somehow reduce its number of chromosome sets to produce haploid gametes; otherwise, the number of chromosome sets will double with every future round of fertilization. Therefore, sexual reproduction requires a nuclear division that reduces the number of chromosome sets by half.
Most animals and plants and many unicellular organisms are diploid and therefore have two sets of chromosomes. In each somatic cell of the organism (all cells of a multicellular organism except the gametes or reproductive cells), the nucleus contains two copies of each chromosome, called homologous chromosomes. Homologous chromosomes are matched pairs containing the same genes in identical locations along their lengths. Diploid organisms inherit one copy of each homologous chromosome from each parent.
Meiosis is the nuclear division that forms haploid cells from diploid cells, and it employs many of the same cellular mechanisms as mitosis. However, as you have learned, mitosis produces daughter cells whose nuclei are genetically identical to the original parent nucleus. In mitosis, both the parent and the daughter nuclei are at the same “ploidy level”—diploid in the case of most multicellular most animals. Plants use mitosis to grow as sporophytes, and to grow and produce eggs and sperm as gametophytes; so they use mitosis for both haploid and diploid cells (as well as for all other ploidies). In meiosis, the starting nucleus is always diploid and the daughter nuclei that result are haploid. To achieve this reduction in chromosome number, meiosis consists of one round of chromosome replication followed by two rounds of nuclear division. Because the events that occur during each of the division stages are analogous to the events of mitosis, the same stage names are assigned. However, because there are two rounds of division, the major process and the stages are designated with a “I” or a “II.” Thus, meiosis I is the first round of meiotic division and consists of prophase I, prometaphase I, and so on. Likewise, Meiosis II (during which the second round of meiotic division takes place) includes prophase II, prometaphase II, and so on.
Meiosis I
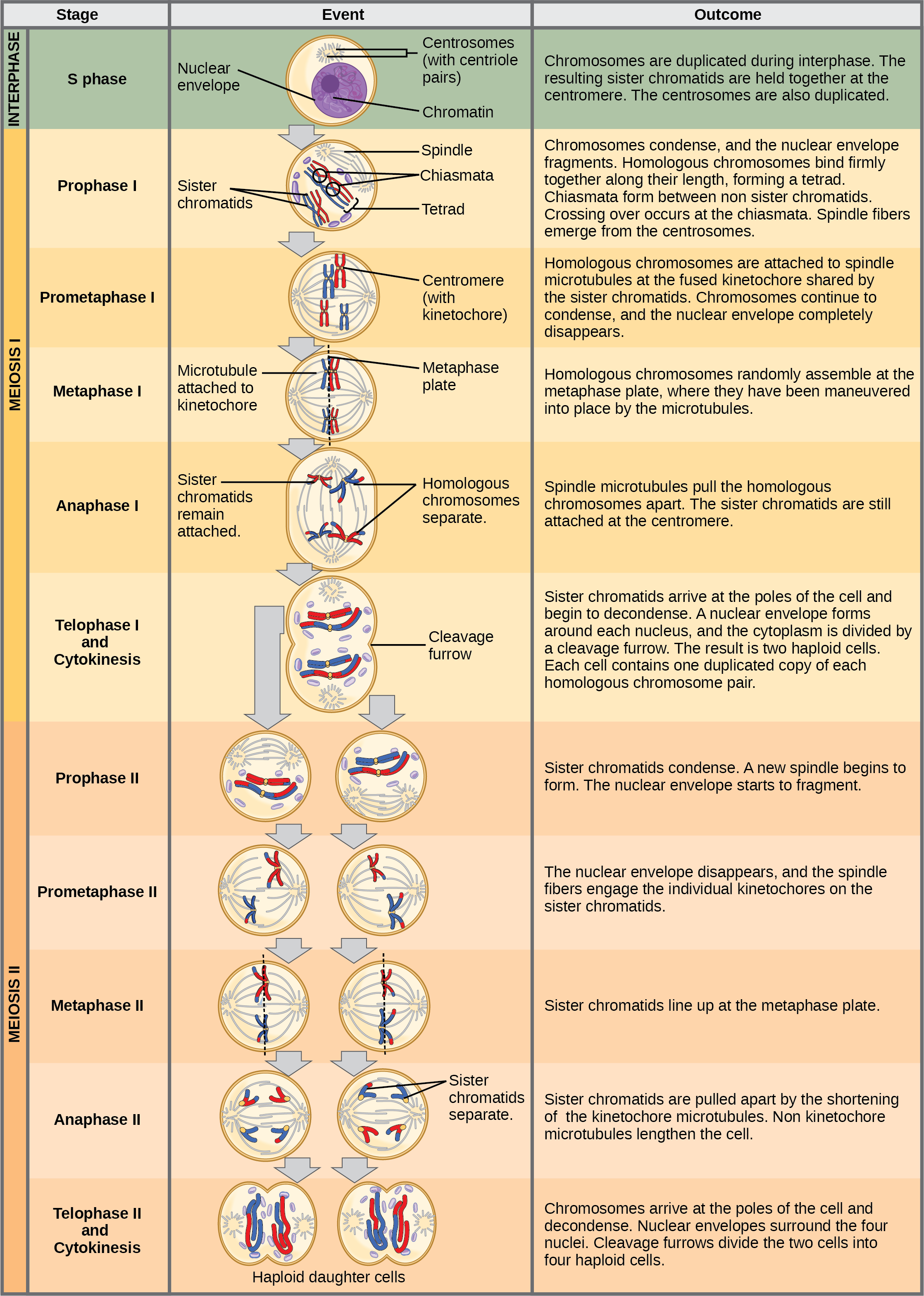
Meiosis is preceded by an interphase consisting of G1, S, and G2 phases, which are nearly identical to the phases preceding mitosis. The G1 phase (the “first gap phase”) is focused on cell growth. During the S phase—the second phase of interphase—the cell copies or replicates the DNA of the chromosomes. Finally, in the G2 phase (the “second gap phase”) the cell undergoes the final preparations for meiosis.
During DNA duplication in the S phase, each chromosome is replicated to produce two identical copies—sister chromatids that are held together at the centromere by cohesin proteins, which hold the chromatids together until anaphase II.
Prophase I
Early in prophase I, before the chromosomes can be seen clearly with a microscope, the homologous chromosomes are attached at their tips to the nuclear envelope by proteins. As the nuclear envelope begins to break down, the proteins associated with homologous chromosomes bring the pair closer together. Recall that in mitosis, homologous chromosomes do not pair together. The synaptonemal complex, a lattice of proteins between the homologous chromosomes, first forms at specific locations and then spreads outward to cover the entire length of the chromosomes. The tight pairing of the homologous chromosomes is called synapsis. In synapsis, the genes on the chromatids of the homologous chromosomes are aligned precisely with each other. The synaptonemal complex supports the exchange of chromosomal segments between homologous nonsister chromatids—a process called crossing over. Crossing over can be observed visually after the exchange as chiasmata (singular = chiasma) ((Figure)).
In humans, even though the X and Y sex chromosomes are not completely homologous (that is, most of their genes differ), there is a small region of homology that allows the X and Y chromosomes to pair up during prophase I. A partial synaptonemal complex develops only between the regions of homology.
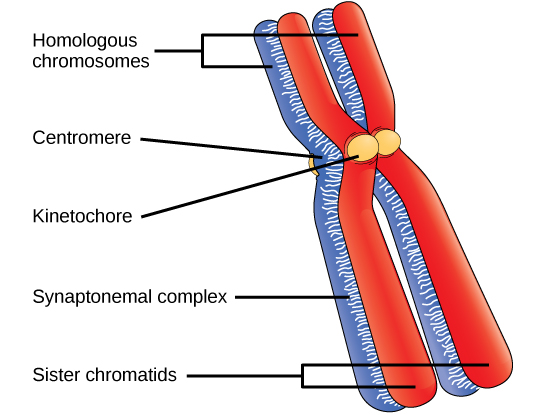
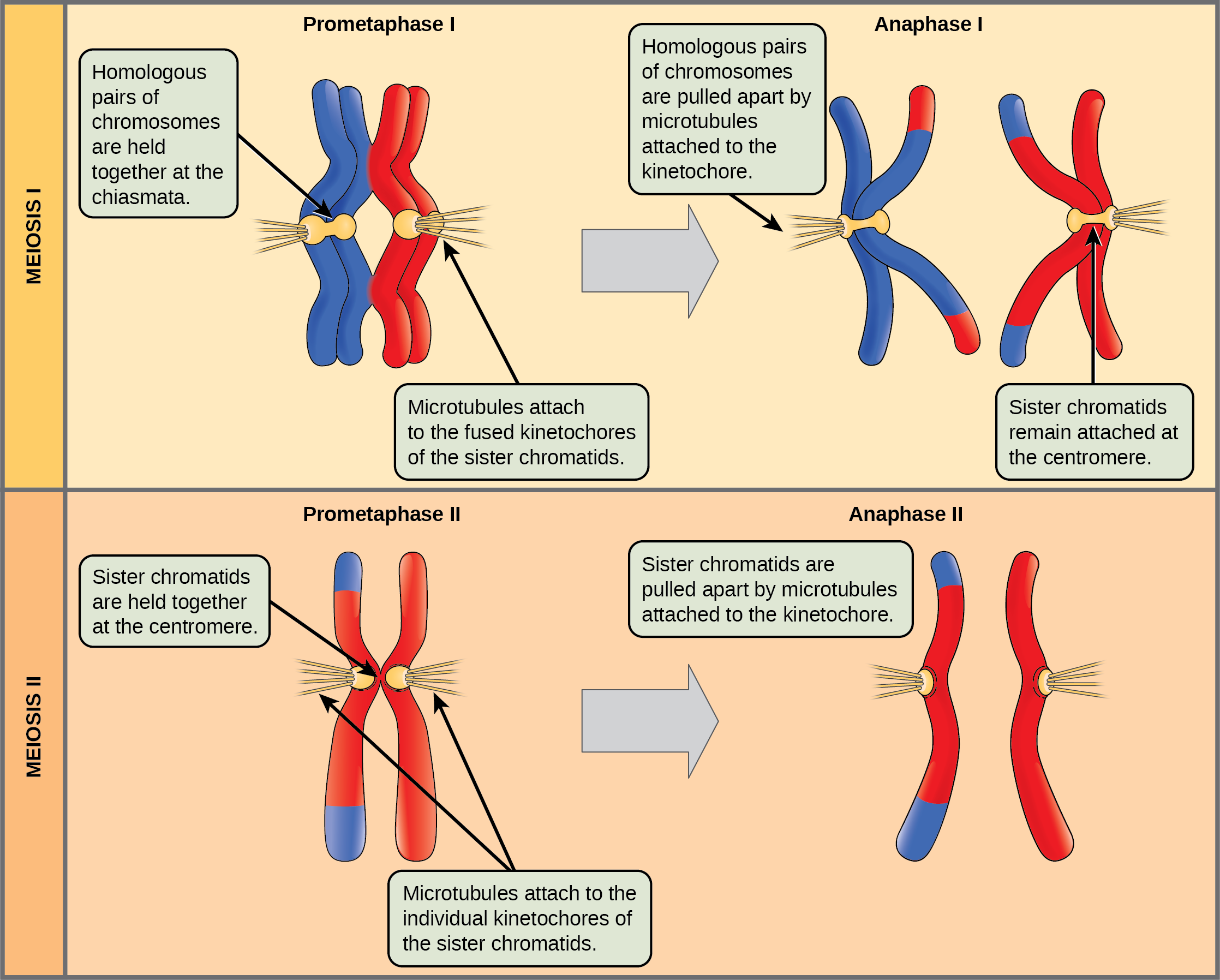
Located at intervals along the synaptonemal complex are large protein assemblies called recombination nodules. These assemblies mark the points of later chiasmata and mediate the multistep process of crossover—or genetic recombination—between the nonsister chromatids. Near the recombination nodule, the double-stranded DNA of each chromatid is cleaved, the cut ends are modified, and a new connection is made between the nonsister chromatids. As prophase I progresses, the synaptonemal complex begins to break down and the chromosomes begin to condense. When the synaptonemal complex is gone, the homologous chromosomes remain attached to each other at the centromere and at chiasmata. The chiasmata remain until anaphase I. The number of chiasmata varies according to the species and the length of the chromosome. There must be at least one chiasma per chromosome for proper separation of homologous chromosomes during meiosis I, but there may be as many as 25. Following crossover, the synaptonemal complex breaks down and the cohesin connection between homologous pairs is removed. At the end of prophase I, the pairs are held together only at the chiasmata ((Figure)). These pairs are called tetrads because the four sister chromatids of each pair of homologous chromosomes are now visible.
The crossover events are the first source of genetic variation in the nuclei produced by meiosis. A single crossover event between homologous nonsister chromatids leads to a reciprocal exchange of equivalent DNA between a maternal chromosome and a paternal chromosome. When a recombinant sister chromatid is moved into a gamete cell it will carry some DNA from one parent and some DNA from the other parent. The recombinant chromatid has a combination of maternal and paternal genes that did not exist before the crossover. Crossover events can occur almost anywhere along the length of the synapsed chromosomes. Different cells undergoing meiosis will therefore produce different recombinant chromatids, with varying combinations of maternal and parental genes. Multiple crossovers in an arm of the chromosome have the same effect, exchanging segments of DNA to produce genetically recombined chromosomes.
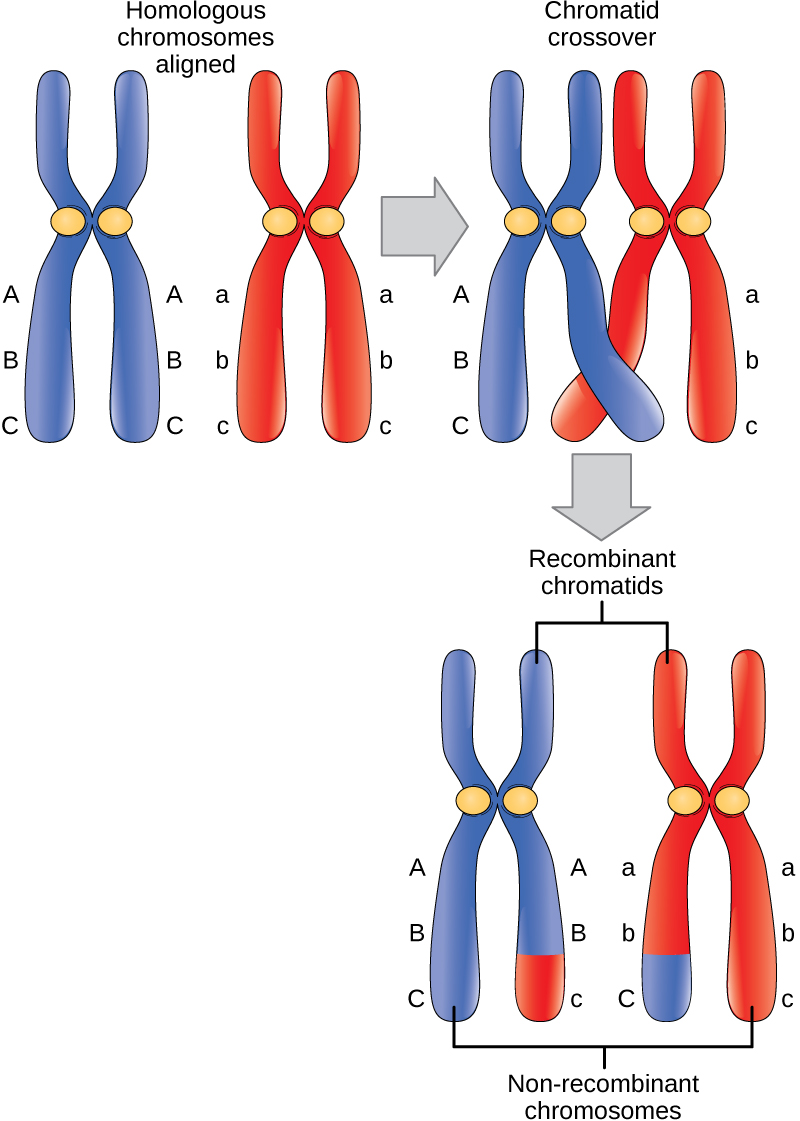
Prometaphase I
The key event in prometaphase I is the attachment of the spindle fiber microtubules to the kinetochore proteins at the centromeres. Kinetochore proteins are multiprotein complexes that bind the centromeres of a chromosome to the microtubules of the mitotic spindle. Microtubules grow from microtubule-organizing centers (MTOCs). In animal cells, MTOCs are centrosomes located at opposite poles of the cell. The microtubules from each pole move toward the middle of the cell and attach to one of the kinetochores of the two fused homologous chromosomes. Each member of the homologous pair attaches to a microtubule extending from opposite poles of the cell so that in the next phase, the microtubules can pull the homologous pair apart. A spindle fiber that has attached to a kinetochore is called a kinetochore microtubule. At the end of prometaphase I, each tetrad is attached to microtubules from both poles, with one homologous chromosome facing each pole. The homologous chromosomes are still held together at the chiasmata. In addition, the nuclear membrane has broken down entirely.
Metaphase I
During metaphase I, the homologous chromosomes are arranged at the metaphase plate—roughly in the midline of the cell, with the kinetochores facing opposite poles. The homologous pairs orient themselves randomly at the equator. For example, if the two homologous members of chromosome 1 are labeled a and b, then the chromosomes could line up a-b or b-a. This is important in determining the genes carried by a gamete, as each will only receive one of the two homologous chromosomes. (Recall that after crossing over takes place, homologous chromosomes are not identical. They contain slight differences in their genetic information, causing each gamete to have a unique genetic makeup.)
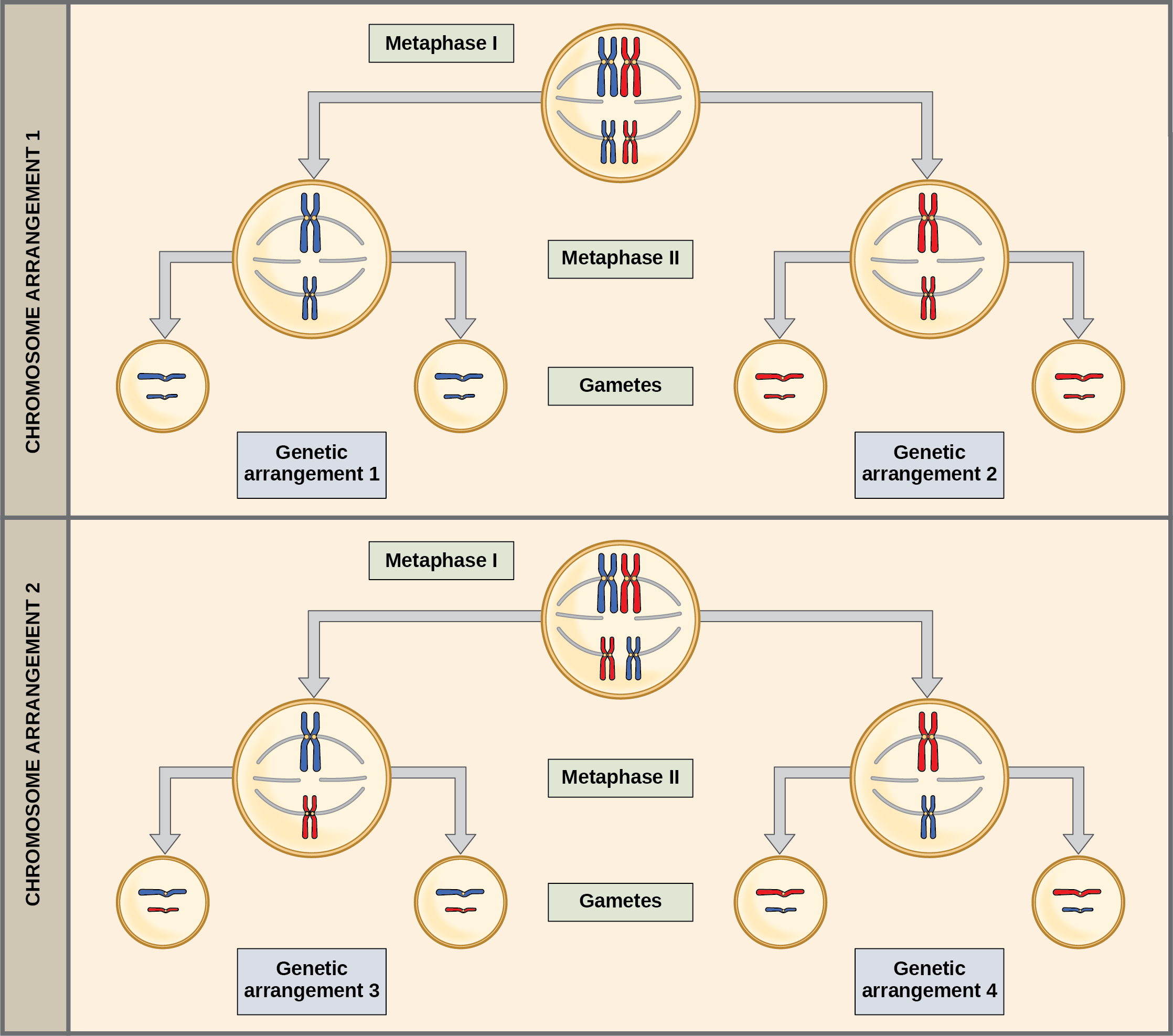
The randomness in the alignment of recombined chromosomes at the metaphase plate, coupled with the crossing over events between nonsister chromatids, are responsible for much of the genetic variation in the offspring. To clarify this further, remember that the homologous chromosomes of a sexually reproducing organism are originally inherited as two separate sets, one from each parent. Using humans as an example, one set of 23 chromosomes is present in the egg donated by the mother. The father provides the other set of 23 chromosomes in the sperm that fertilizes the egg. Every cell of the multicellular offspring has copies of the original two sets of homologous chromosomes. In prophase I of meiosis, the homologous chromosomes form the tetrads. In metaphase I, these pairs line up at the midway point between the two poles of the cell to form the metaphase plate. Because there is an equal chance that a microtubule fiber will encounter a maternally or paternally inherited chromosome, the arrangement of the tetrads at the metaphase plate is random. Thus, any maternally inherited chromosome may face either pole. Likewise, any paternally inherited chromosome may also face either pole. The orientation of each tetrad is independent of the orientation of the other 22 tetrads.
This event—the random (or independent) assortment of homologous chromosomes at the metaphase plate—is the second mechanism that introduces variation into the gametes or spores. In each cell that undergoes meiosis, the arrangement of the tetrads is different. The number of variations is dependent on the number of chromosomes making up a set. There are two possibilities for orientation at the metaphase plate; the possible number of alignments therefore equals 2n in a diploid cell, where n is the number of chromosomes per haploid set. Humans have 23 chromosome pairs, which results in over eight million (223) possible genetically-distinct gametes just from the random alignment of chromosomes at the metaphase plate. This number does not include the variability that was previously produced by crossing over between the nonsister chromatids. Given these two mechanisms, it is highly unlikely that any two haploid cells resulting from meiosis will have the same genetic composition ((Figure)).
To summarize, meiosis I creates genetically diverse gametes in two ways. First, during prophase I, crossover events between the nonsister chromatids of each homologous pair of chromosomes generate recombinant chromatids with new combinations of maternal and paternal genes. Second, the random assortment of tetrads on the metaphase plate produces unique combinations of maternal and paternal chromosomes that will make their way into the gametes.
Anaphase I
In anaphase I, the microtubules pull the linked chromosomes apart. The sister chromatids remain tightly bound together at the centromere. The chiasmata are broken in anaphase I as the microtubules attached to the fused kinetochores pull the homologous chromosomes apart ((Figure)).
Telophase I and Cytokinesis
In telophase, the separated chromosomes arrive at opposite poles. The remainder of the typical telophase events may or may not occur, depending on the species. In some organisms, the chromosomes “decondense” and nuclear envelopes form around the separated sets of chromatids produced during telophase I. In other organisms, cytokinesis—the physical separation of the cytoplasmic components into two daughter cells—occurs without reformation of the nuclei. In nearly all species of animals and some fungi, cytokinesis separates the cell contents via a cleavage furrow (constriction of the actin ring that leads to cytoplasmic division). In plants, a cell plate is formed during cell cytokinesis by Golgi vesicles fusing at the metaphase plate. This cell plate will ultimately lead to the formation of cell walls that separate the two daughter cells.
Two haploid cells are the result of the first meiotic division of a diploid cell. The cells are haploid because at each pole, there is just one of each pair of the homologous chromosomes. Therefore, only one full set of the chromosomes is present. This is why the cells are considered haploid—there is only one chromosome set, even though each chromosome still consists of two sister chromatids. Recall that sister chromatids are merely duplicates of one of the two homologous chromosomes (except for changes that occurred during crossing over). In meiosis II, these two sister chromatids will separate, creating four haploid daughter cells.
Link to Learning
Review the process of meiosis, observing how chromosomes align and migrate, at Meiosis: An Interactive Animation.
Meiosis II
In some species, cells enter a brief interphase, or interkinesis, before entering meiosis II. Interkinesis lacks an S phase, so chromosomes are not duplicated. The two cells produced in meiosis I go through the events of meiosis II in synchrony. During meiosis II, the sister chromatids within the two daughter cells separate, forming four new haploid gametes. The mechanics of meiosis II are similar to mitosis, except that each dividing cell has only one set of homologous chromosomes, each with two chromatids. Therefore, each cell has half the number of sister chromatids to separate out as a diploid cell undergoing mitosis. In terms of chromosomal content, cells at the start of meiosis II are similar to haploid cells in G2, preparing to undergo mitosis.
Prophase II
If the chromosomes decondensed in telophase I, they condense again. If nuclear envelopes were formed, they fragment into vesicles. The MTOCs that were duplicated during interkinesis move away from each other toward opposite poles, and new spindles are formed.
Prometaphase II
The nuclear envelopes are completely broken down, and the spindle is fully formed. Each sister chromatid forms an individual kinetochore that attaches to microtubules from opposite poles.
Metaphase II
The sister chromatids are maximally condensed and aligned at the equator of the cell.
Anaphase II
The sister chromatids are pulled apart by the kinetochore microtubules and move toward opposite poles. Nonkinetochore microtubules elongate the cell.
Telophase II and Cytokinesis
The chromosomes arrive at opposite poles and begin to decondense. Nuclear envelopes form around the chromosomes. If the parent cell was diploid, as is most commonly the case, then cytokinesis now separates the two cells into four unique haploid cells. The cells produced are genetically unique because of the random assortment of paternal and maternal homologs and because of the recombination of maternal and paternal segments of chromosomes (with their sets of genes) that occurs during crossover. The entire process of meiosis is outlined in (Figure).
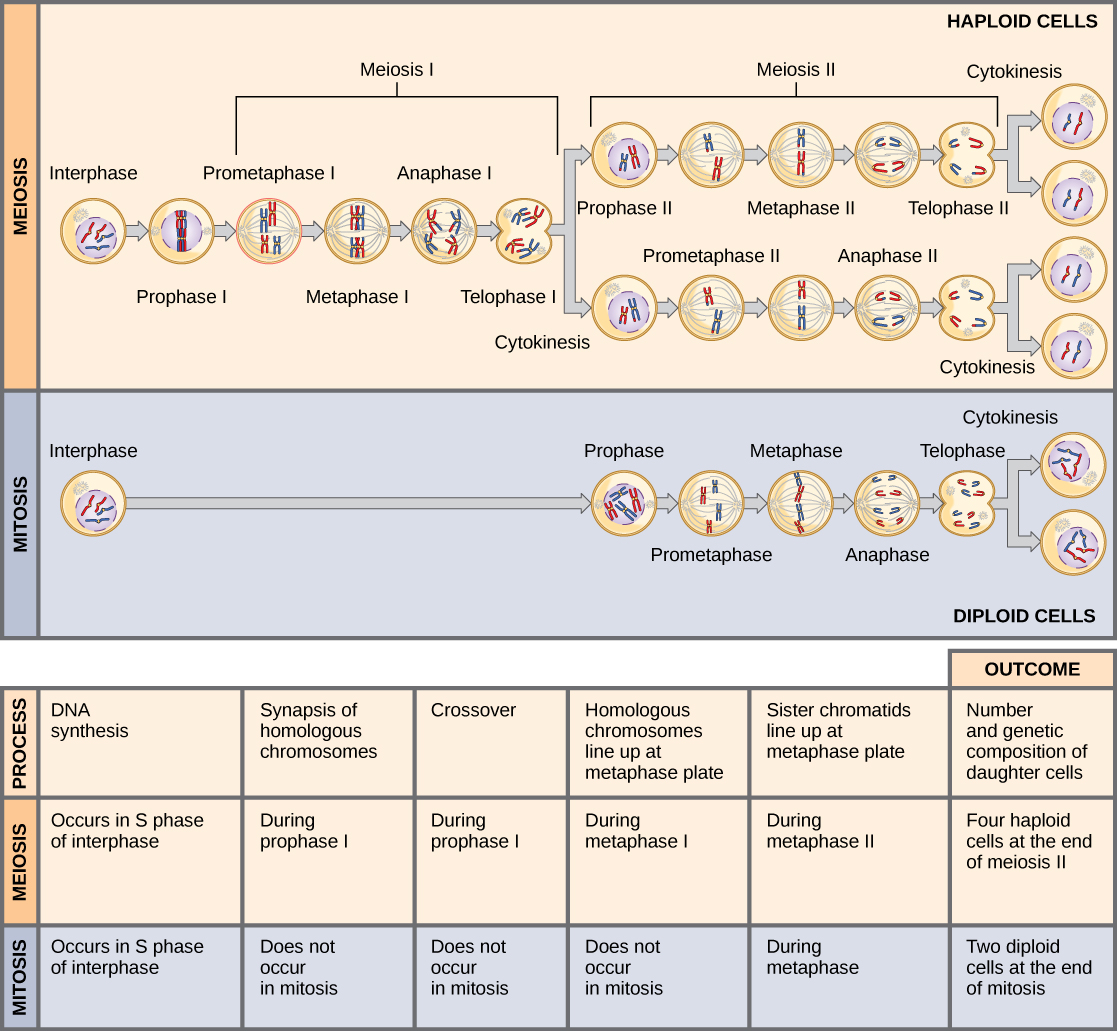
Comparing Meiosis and Mitosis
Mitosis and meiosis are both forms of division of the nucleus in eukaryotic cells. They share some similarities, but also exhibit a number of important and distinct differences that lead to very different outcomes ((Figure)). Mitosis is a single nuclear division that results in two nuclei that are usually partitioned into two new cells. The nuclei resulting from a mitotic division are genetically identical to the original nucleus. They have the same number of sets of chromosomes: one set in the case of haploid cells and two sets in the case of diploid cells. In contrast, meiosis consists of two nuclear divisions resulting in four nuclei that are usually partitioned into four new, genetically distinct cells. The four nuclei produced during meiosis are not genetically identical, and they contain one chromosome set only. This is half the number of chromosome sets in the original cell, which is diploid.
The main differences between mitosis and meiosis occur in meiosis I, which is a very different nuclear division than mitosis. In meiosis I, the homologous chromosome pairs physically meet and are bound together with the synaptonemal complex. Following this, the chromosomes develop chiasmata and undergo crossover between nonsister chromatids. In the end, the chromosomes line up along the metaphase plate as tetrads—with kinetochore fibers from opposite spindle poles attached to each kinetochore of a homolog to form a tetrad. All of these events occur only in meiosis I.
When the chiasmata resolve and the tetrad is broken up with the homologs moving to one pole or another, the ploidy level—the number of sets of chromosomes in each future nucleus—has been reduced from two to one. For this reason, meiosis I is referred to as a reductional division. There is no such reduction in ploidy level during mitosis.
Meiosis II is analogous to a mitotic division. In this case, the duplicated chromosomes (only one set of them) line up on the metaphase plate with divided kinetochores attached to kinetochore fibers from opposite poles. During anaphase II, as in mitotic anaphase, the kinetochores divide and one sister chromatid—now referred to as a chromosome—is pulled to one pole while the other sister chromatid is pulled to the other pole. If it were not for the fact that there had been crossover, the two products of each individual meiosis II division would be identical (as in mitosis). Instead, they are different because there has always been at least one crossover per chromosome. Meiosis II is not a reduction division because although there are fewer copies of the genome in the resulting cells, there is still one set of chromosomes, as there was at the end of meiosis I.
Evolution Connection
The Mystery of the Evolution of MeiosisSome characteristics of organisms are so widespread and fundamental that it is sometimes difficult to remember that they evolved like other simple traits. Meiosis is such an extraordinarily complex series of cellular events that biologists have had trouble testing hypotheses concerning how it may have evolved. Although meiosis is inextricably entwined with sexual reproduction and its advantages and disadvantages, it is important to separate the questions of the evolution of meiosis and the evolution of sex, because early meiosis may have been advantageous for different reasons than it is now. Thinking outside the box and imagining what the early benefits from meiosis might have been is one approach to uncovering how it may have evolved.
Meiosis and mitosis share obvious cellular processes, and it makes sense that meiosis evolved from mitosis. The difficulty lies in the clear differences between meiosis I and mitosis. Adam Wilkins and Robin Holliday1 summarized the unique events that needed to occur for the evolution of meiosis from mitosis. These steps are homologous chromosome pairing and synapsis, crossover exchanges, sister chromatids remaining attached during anaphase, and suppression of DNA replication in interphase. They argue that the first step is the hardest and most important and that understanding how it evolved would make the evolutionary process clearer. They suggest genetic experiments that might shed light on the evolution of synapsis.
There are other approaches to understanding the evolution of meiosis in progress. Different forms of meiosis exist in single-celled protists. Some appear to be simpler or more “primitive” forms of meiosis. Comparing the meiotic divisions of different protists may shed light on the evolution of meiosis. Marilee Ramesh and colleagues2 compared the genes involved in meiosis in protists to understand when and where meiosis might have evolved. Although research is still ongoing, recent scholarship into meiosis in protists suggests that some aspects of meiosis may have evolved later than others. This kind of genetic comparison can tell us what aspects of meiosis are the oldest and what cellular processes they may have borrowed from in earlier cells.
Link to Learning
Click through the steps of this interactive animation to compare the meiotic process of cell division to that of mitosis at How Cells Divide.
Section Summary
Sexual reproduction requires that organisms produce cells that can fuse during fertilization to produce offspring. In most animals, meiosis is used to produce haploid eggs and sperm from diploid parent cells so that the fusion of an egg and sperm produces a diploid zygote. As with mitosis, DNA replication occurs prior to meiosis during the S-phase of the cell cycle so that each chromosome becomes a pair of sister chromatids. In meiosis, there are two rounds of nuclear division resulting in four nuclei and usually four daughter cells, each with half the number of chromosomes as the parent cell. The first division separates homologs, and the second—like mitosis—separates chromatids into individual chromosomes. Meiosis generates variation in the daughter nuclei during crossover in prophase I as well as during the random alignment of tetrads at metaphase I. The cells that are produced by meiosis are genetically unique.
Meiosis and mitosis share similar processes, but have distinct outcomes. Mitotic divisions are single nuclear divisions that produce genetically identical daughter nuclei (i.e., each daughter nucleus has the same number of chromosome sets as the original cell). In contrast, meiotic divisions include two nuclear divisions that ultimately produce four genetically different daughter nuclei that have only one chromosome set (instead of the two sets of chromosomes in the parent cell). The main differences between the two nuclear division processes take place during the first division of meiosis: homologous chromosomes pair, crossover, and exchange homologous nonsister chromatid segments. The homologous chromosomes separate into different nuclei during meiosis I, causing a reduction of ploidy level in the first division. The second division of meiosis is similar to a mitotic division, except that the daughter cells do not contain identical genomes because of crossover and chromosome recombination in prophase I.
Footnotes
- 1 Adam S. Wilkins and Robin Holliday, “The Evolution of Meiosis from Mitosis,” Genetics 181 (2009): 3–12.
- 2 Marilee A. Ramesh, Shehre-Banoo Malik and John M. Logsdon, Jr, “A Phylogenetic Inventory of Meiotic Genes: Evidence for Sex in Giardia and an Early Eukaryotic Origin of Meiosis,” Current Biology 15 (2005):185–91.
Glossary
- chiasmata
- (singular, chiasma) the structure that forms at the crossover points after genetic material is exchanged
- cohesin
- proteins that form a complex that seals sister chromatids together at their centromeres until anaphase II of meiosis
- crossover
- exchange of genetic material between nonsister chromatids resulting in chromosomes that incorporate genes from both parents of the organism
- fertilization
- union of two haploid cells from two individual organisms
- interkinesis
- (also, interphase II) brief period of rest between meiosis I and meiosis II
- meiosis
- a nuclear division process that results in four haploid cells
- meiosis I
- first round of meiotic cell division; referred to as reduction division because the ploidy level is reduced from diploid to haploid
- meiosis II
- second round of meiotic cell division following meiosis I; sister chromatids are separated into individual chromosomes, and the result is four unique haploid cells
- recombination nodules
- protein assemblies formed on the synaptonemal complex that mark the points of crossover events and mediate the multistep process of genetic recombination between nonsister chromatids
- reduction division
- nuclear division that produces daughter nuclei each having one-half as many chromosome sets as the parental nucleus; meiosis I is a reduction division
- somatic cell
- all the cells of a multicellular organism except the gametes or reproductive cells
- spore
- haploid cell that can produce a haploid multicellular organism or can fuse with another spore to form a diploid cell
- synapsis
- formation of a close association between homologous chromosomes during prophase I
- synaptonemal complex
- protein lattice that forms between homologous chromosomes during prophase I, supporting crossover
- tetrad
- two duplicated homologous chromosomes (four chromatids) bound together by chiasmata during prophase I

